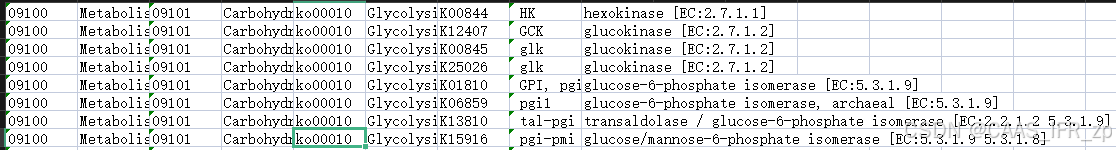
首先进入KEGG BRITE: KEGG Orthology (KO)
下载json文件
用python处理一下
import json
import re
import os
os.chdir("C:/Users/fordata/Downloads/")
with open("ko00001.json","r") as f:
fj = f.read()
kojson = json.loads(fj)
with open("newKegg.tsv", "w") as k:
for i in kojson['children']:
ii = i['name'].replace(" ", "\t", 1)
for j in i['children']:
jj = j['name'].replace(" ", "\t", 1)
for m in j['children']:
if re.findall(r"ko\d{5}", m['name']):
mm = "ko" + m['name'].replace(" ", "\t", 1)
else:
mm = m['name'].replace(" ", "\t", 1)
try:
for n in m['children']:
if ";" in n['name']:
nn = n['name'].replace(" ", "\t", 1).replace("; ", "\t", 1)
else:
nn = n['name'].replace(" ", "\t \t", 1)
k.write(ii + "\t" + jj + "\t" + mm + "\t" + nn + "\n")
except:
nn = " \t \t "
k.write(ii+"\t"+jj+"\t"+mm+"\t"+nn+"\n")得到结果
写个代码看看把keggKO和tpm关联起来
#! /usr/bin/env python
#########################################################
# mix eggnog(kegg) result with tpm
# written by PeiZhong in IFR of CAAS
import argparse
import pandas as pd
# Parse command-line arguments
parser = argparse.ArgumentParser(description='Mix eggnog(kegg) result with TPM')
parser.add_argument('--result', "-r", required=True, help='Path to eggnog result file')
parser.add_argument('--tpm', "-t", required=True, help='Path to TPM table file')
parser.add_argument('--out', "-o", required=True, help='Path to output file')
args = parser.parse_args()
# Step 1: Read input files
print("Reading input files")
# Read dbcan result
df_result = {}
df_kegg = set() # Use a set to store unique CAZy families
with open(args.result, "r") as f:
for line in f:
if "#" not in line:
protein_id = line.split("\t")[0]
kegg_str = line.split("\t")[11]
if "-" != kegg_str:
df_result[protein_id] = kegg_str
# Extract CAZy families and remove duplicates
families = set(entry.split(":")[1].strip() for entry in kegg_str.split(','))
df_kegg.update(families) # Add unique families to the global set
# Read TPM file
df_tpm = pd.read_csv(args.tpm, sep='\t')
# Step 2: Process dbcan results and calculate TPM sums for each sample
print("Processing dbcan results and calculating TPM sums for each sample")
# Initialize a dictionary to store TPM sums for each CAZy family and sample
kegg_tpm_sums = {ko: {sample: 0.0 for sample in df_tpm.columns[1:]} for ko in df_kegg}
# Convert TPM table to a dictionary for faster lookup
tpm_dict = df_tpm.set_index(df_tpm.columns[0]).to_dict(orient='index')
# Process each protein in the dbcan result
for protein_id, kegg_str in df_result.items():
# Convert protein ID to gene ID by removing trailing "_number"
if "_" in protein_id:
gene_id = protein_id.rsplit("_", 1)[0] # Split from right on the last "_"
else:
print(f"Warning: Protein ID {protein_id} has no underscore, using as gene ID")
gene_id = protein_id
# Get TPM values for this gene
if gene_id not in tpm_dict:
print(f"Warning: No TPM values found for {gene_id} (protein {protein_id})")
continue
tpm_values = tpm_dict[gene_id]
# Extract unique CAZy families for this protein
families = set(entry.split(':')[1].strip() for entry in kegg_str.split(','))
# Update TPM sums for each unique CAZy family
for family in families:
if family in kegg_tpm_sums:
for sample in df_tpm.columns[1:]:
kegg_tpm_sums[family][sample] += tpm_values[sample]
else:
# Dynamically add new CAZy families
kegg_tpm_sums[family] = {sample: tpm_values[sample] for sample in df_tpm.columns[1:]}
# Create and save output DataFrame
output_df = pd.DataFrame.from_dict(kegg_tpm_sums, orient='index')
output_df.index.name = 'CAZy_Family'
output_df.to_csv(args.out, sep='\t', float_format='%.2f') # Round to 2 decimal places
print(f"Results saved to {args.out}")得到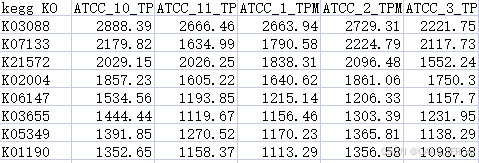
kegg的对应level,在excel钟使用vlookup函数对应即可