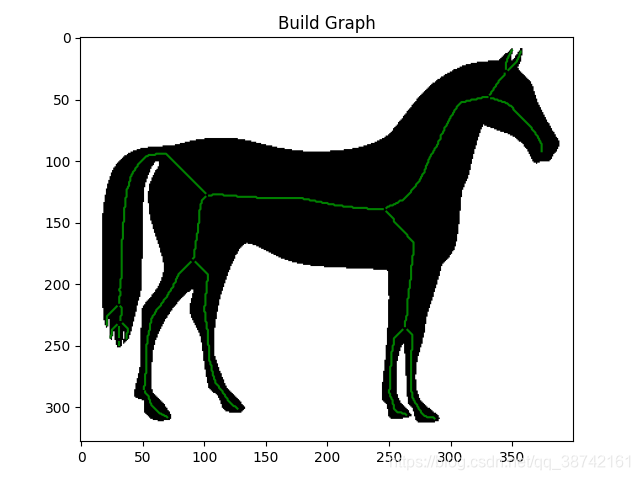
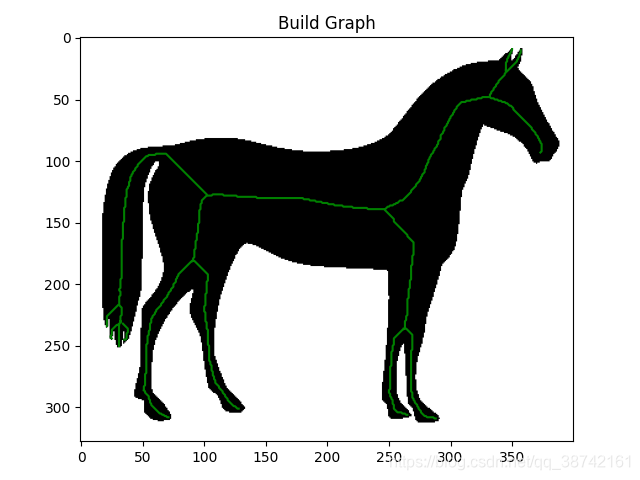
路网分割后得到region,提取骨架得到centerline,之后需要进行矢量化得到结点和边,进而转化成geojson格式进行生产。
本文对矢量化函数库sknw源码进行研读,并改进源码使结点和边之间紧密连接。
一、骨架提取并矢量化demo
from skimage.morphology import skeletonize
from skimage import data
import sknw
import numpy as np
import matplotlib.pyplot as plt
# 骨架提取
img = data.horse()
ske = skeletonize(~img).astype(np.uint16)
# 矢量化调用函数
graph = sknw.build_sknw(ske)
# draw image
plt.imshow(img, cmap='gray')
# draw edges by pts
for (s, e) in graph.edges():
ps = graph[s][e]['pts']
plt.plot(ps[:, 1], ps[:, 0], 'green')
# draw node by o
# node, nodes = graph._node, graph.nodes()
# ps = np.array([node[i]['o'] for i in nodes])
# plt.plot(ps[:, 1], ps[:, 0], 'r.')
# title and show
plt.title('Build Graph')
plt.show()
# plt.savefig('pc.png')二、sknw源码研读:
①做一个像素的buffer,以免原图边界处找不到3*3邻域。
②将二值化图像进行结点映射,背景0,边1,结点2
③结点域提取,相邻结点组成一个结点域,对每个结点域进行索引编码,从10开始,依次递增。为了避免与映射012混淆,10代表第0个结点。
④遍历结点作为入口,遍历邻域寻找线,如果邻域内两次找到结点,则已遍历到的线作为这两个结点的连线。
⑤结点域找中心点
import numpy as np
from numba import jit
import networkx as nx
import matplotlib.pyplot as plt
# get neighbors d index
def neighbors(shape):
"""
找出3*3大小的邻域,并压缩至向量的形式表示
"""
dim = len(shape)
block = np.ones([3]*dim)
block[tuple([1]*dim)] = 0
idx = np.where(block>0)
idx = np.array(idx, dtype=np.uint8).T
idx = np.array(idx-[1]*dim)
acc = np.cumprod((1,)+shape[::-1][:-1])
return np.dot(idx, acc[::-1])
@jit # my markimport m
def mark(img): # mark the array use (0, 1, 2)
"""
将二值化的骨架图按照背景、线、结点的形式映射到0,1,2
"""
nbs = neighbors(img.shape)
H,W = img.shape
img = img.ravel()
for p in range(len(img)):
if img[p]==0:continue
s = 0
for dp in nbs:
if img[p+dp]!=0:s+=1
if s==2:img[p]=1
else:img[p]=2
# image = np.zeros((H,W))
# for i in range(len(img)):
# image[i//W,i-i//W*W] = img[i]
# tmp = image[200:251,:51]
# plt.imshow(tmp,cmap="gray")
# plt.show()
@jit # trans index to r, c...
def idx2rc(idx, acc):
"""
将一维向量形式的坐标映射到二维图像坐标
"""
rst = np.zeros((len(idx), len(acc)), dtype=np.int16)
for i in range(len(idx)):
for j in range(len(acc)):
rst[i,j] = idx[i]//acc[j]
idx[i] -= rst[i,j]*acc[j]
rst -= 1
return rst
@jit # fill a node (may be two or more points)
def fill(img, p, num, nbs, acc, buf):
"""
cur 当前遍历的结点,s 当前存储结点,该循环用以遍历所有相邻(8邻域)的node结点。
return 二维list:以p点为中心进行拓展,找出包含p的所有密闭链接的结点。形式:[[node1_x,node2_y]...]
"""
#back = 2
back = img[p]
img[p] = num
#buf存储idx
buf[0] = p
cur = 0; s = 1;
while True:
p = buf[cur]
for dp in nbs:
cp = p+dp
if img[cp]==back:
img[cp] = num
buf[s] = cp
s+=1
cur += 1
if cur==s:break
return idx2rc(buf[:s], acc)
@jit # trace the edge and use a buffer, then buf.copy, if use [] numba not works
def trace(img, p, nbs, acc, buf):
"""
c1 头结点索引, c2 尾结点索引, 注意有着先后(小大)顺序,顺序不能乱,否则后续连线出现飞线
newp 存储线上要遍历的下一个点。
修改方法:我们将头尾结点添加至线的范围内,这样可以连接结点域内部结点间的线。从而生成封闭的拓扑。
"""
c1 = 0; c2 = 0;
newp = 0
cur = 0
while True:
buf[cur] = p
img[p] = 0
cur += 1
for dp in nbs:
cp = p + dp
if img[cp] >= 10:
if c1==0:
c1=img[cp]
#add
# c1_p = cp
else:
c2 = img[cp]
#add
# c2_p = cp
if img[cp] == 1:
newp = cp
p = newp
if c2!=0:break
# #add
# buf = np.insert(buf,0,c1_p)
# #add
# buf[cur+1] = c2_p
# #add
# cur += 2
return (c1-10, c2-10, idx2rc(buf[:cur], acc))
@jit # parse the image then get the nodes and edges
def parse_struc(img):
#img.shape H*W
nbs = neighbors(img.shape)
#acc: (W,1)
acc = np.cumprod((1,)+img.shape[::-1][:-1])[::-1]
img = img.ravel()
#pts: 结点索引,平铺后
pts = np.array(np.where(img==2))[0]
buf = np.zeros(131072, dtype=np.int64)
#num 结点索引,以10开始,为了避免mark(0 1 2)的干扰,所以从10开始代表第0个结点域(注意是一片连续的区域)。每存储一个结点域num+1。
num = 10
nodes = []
for p in pts:
if img[p] == 2:
nds = fill(img, p, num, nbs, acc, buf)
num += 1
nodes.append(nds)
edges = []
for p in pts:
for dp in nbs:
if img[p+dp]==1:
edge = trace(img, p+dp, nbs, acc, buf)
edges.append(edge)
return nodes, edges
# use nodes and edges build a networkx graph
def build_graph(nodes, edges, multi=False):
graph = nx.MultiGraph() if multi else nx.Graph()
for i in range(len(nodes)):
graph.add_node(i, pts=nodes[i], o=nodes[i].mean(axis=0))
for s,e,pts in edges:
l = np.linalg.norm(pts[1:]-pts[:-1], axis=1).sum()
graph.add_edge(s,e, pts=pts, weight=l)
return graph
def buffer(ske):
#扩充一个单位像素的边,以便对原图中每个像素进行八邻域查找
buf = np.zeros(tuple(np.array(ske.shape)+2), dtype=np.uint16)
buf[tuple([slice(1,-1)]*buf.ndim)] = ske
return buf
def build_sknw(ske, multi=False):
buf = buffer(ske)
mark(buf)
nodes, edges = parse_struc(buf)
return build_graph(nodes, edges, multi)
# draw the graph
def draw_graph(img, graph, cn=255, ce=128):
acc = np.cumprod((1,)+img.shape[::-1][:-1])[::-1]
img = img.ravel()
for idx in graph.nodes():
pts = graph.node[idx]['pts']
img[np.dot(pts, acc)] = cn
for (s, e) in graph.edges():
eds = graph[s][e]
for i in eds:
pts = eds[i]['pts']
img[np.dot(pts, acc)] = ce
if __name__ == '__main__':
g = nx.MultiGraph()
g.add_nodes_from([1,2,3,4,5])
g.add_edges_from([(1,2),(1,3),(2,3),(4,5),(5,4)])
print(g.nodes())
print(g.edges())
a = g.subgraph(1)
print('d')
print(a)
print('d')
最后输出每个结点域的中心点和线。但是存在结点与线分离的情况。如下图所示:
三、添加代码:结点与边相连
分析不相连的原因:结点域以结点中心区域表示,故其它结点不会显示,于是存在断线。我的思路是从线的端点开始,向外遍历并入非结点中心的结点作为线的扩充点即可。注意线的顺序,因为线的存储是有序的,头结点和尾结点的顺序正好相反。添加代码如下:
#add
def join_nodes(graph):
node, nodes = graph._node, graph.nodes()
center_node = np.array([node[i]['o'] for i in nodes])
all_nodes = np.array([node[i]['pts'] for i in nodes])
for (s, e) in graph.edges():
ps = graph[s][e]['pts']
s_center_node = center_node[s]
e_center_node = center_node[e]
s_all_nodes = all_nodes[s]
e_all_nodes = all_nodes[e]
s_line_point = ps[0]
e_line_point = ps[-1]
#线长度为一的不进行扩展,以免后续清洗不掉
if len(ps)==1:
continue
if len(s_all_nodes)==1:
graph[s][e]['pts'] = np.vstack((s_center_node,graph[s][e]['pts']))
else:
bbox = [min(s_center_node[0],s_line_point[0]),max(s_center_node[0],s_line_point[0]),
min(s_center_node[1],s_line_point[1]),max(s_center_node[1],s_line_point[1])]
s_crop_nodes = [i for i in s_all_nodes if i[0]>=bbox[0] and i[0]<=bbox[1] and i[1]>=bbox[2] and i[1]<=bbox[3]][::-1]
for i in s_crop_nodes:
graph[s][e]['pts'] = np.vstack((np.array(i),graph[s][e]['pts']))
if len(e_all_nodes)==1:
graph[s][e]['pts'] = np.vstack((graph[s][e]['pts'],e_center_node))
else:
bbox = [min(e_center_node[0],e_line_point[0]),max(e_center_node[0],e_line_point[0]),
min(e_center_node[1],e_line_point[1]),max(e_center_node[1],e_line_point[1]),]
e_crop_nodes = [i for i in e_all_nodes if i[0]>=bbox[0] and i[0]<=bbox[1] and i[1]>=bbox[2] and i[1]<=bbox[3]][::-1]
for i in e_crop_nodes:
graph[s][e]['pts'] = np.vstack((graph[s][e]['pts'],np.array(i)))
return graph
graph = join_nodes(graph)后处理改进结果: