文章目录
BDF 转 MAT 讲解文档
一、概述
BDF(BioSemi Data Format)是一种常用的生物医学信号数据格式,而MAT是MATLAB软件的数据格式。在进行生物医学信号分析时,有时需要将BDF文件转换为MAT文件,以便在MATLAB环境中进行进一步处理和分析。本文将介绍两种将BDF文件转换为MAT文件的方法:使用自定义函数readbdf和利用EEGLAB界面操作。
二、使用自定义函数readbdf进行转换
(一)函数简介
readbdf是一个专门用于读取BDF文件并将其转换为MAT文件的函数。该函数能够解析BDF文件中的信号数据、采样率、通道信息等关键数据,并将其存储为MATLAB可识别的变量格式,方便后续处理。
(二)使用步骤
- 准备函数文件
确保你已经获得了readbdf函数的源代码文件,通常为.m格式。将该文件保存在MATLAB的工作路径下,或者添加其所在路径到MATLAB的搜索路径中,以便MATLAB能够找到并调用该函数。
- 调用函数读取BDF文件
在MATLAB命令窗口或脚本文件中,使用以下命令调用readbdf函数:
function EEG = readbdfdata(filename, pathname)
%
% Syntax: EEG = readbdfdata(pathname)
%
%
% Inputs:
% filename:
% pathname: path to data files
% Outputs:
% EEG data structure
%
% Example:
%
%
%
% Subfunctions: read_bdf, readLowLevel, readEvents
% MAT-files required: none
%
%
% Versions:
% v0.1: 2017-09-27, orignal
%
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
%find the index of data file
index = [];
if ~iscell(filename)
filename = {filename};
end
for i = 1:length(filename)
if length(filename{i}) >= 4 %length('data')==4
if isequal(filename{i}(1:4), 'data')
index = [index, i];
end
end
end
datafilelength = length(index);
%read data files
if datafilelength >0
%add information for eeglab structure
EEG.setname = 'BDF file';
EEG.filename = char(filename);
EEG.filepath = pathname;
EEG.subject = '';
EEG.group = '';
EEG.condition = '';
EEG.session = [];
EEG.comments = ['Original files:' pathname];
EEG.icaact = [];
EEG.icawinv = [];
EEG.icasphere = [];
EEG.icaweights = [];
EEG.icachansind = [];
chaninfo.plotrad = [];
chaninfo.shrink = [];
chaninfo.nosedir = '+X';
chaninfo.icachansind = [];
EEG.chaninfo = chaninfo;
EEG.ref = 'common';
EEG.specdata = [];
EEG.specicaact = [];
EEG.splinefile = '';
EEG.icasplinefile = '';
EEG.dipfit = [];
EEG.xmin = 0;
EEG.xmax = 0;
EEG.pnts = 0;
T0 = [];
EEG.urevent = [];
EEG.eventdescription = {};
EEG.epoch = [];
EEG.epochdescription = {};
EEG.data = [];
Data = {};
datapnts = [];
for i = 1:datafilelength
datafilename = [pathname filename{index(i)}];
%read header
hdr = read_bdf(datafilename);
EEG.srate = hdr.Fs;
EEG.nbchan = hdr.nChans;
datapnts = [datapnts,hdr.nSamples];
EEG.pnts = EEG.pnts+hdr.nSamples;
EEG.chanlocs= hdr.chanlocs;
EEG.trials = hdr.nTrials;
EEG.xmax = EEG.xmax+EEG.pnts/EEG.srate; %in seconds
T0 = [T0; hdr.T0];
%compatible with older version data file which has event channel
if hdr.Annotation == 1
evt = hdr.event;
evt = cell2mat(evt);
EEG.event = struct([]);
if isstruct(evt)
for kk = 1:size(evt,2)
event = struct('type',evt(kk).eventvalue,'latency',evt(kk).offset_in_sec*EEG.srate);
EEG.event = [EEG.event;event];
end
end
end
if datafilelength == 1
EEG.data = read_bdf(datafilename,hdr,1,hdr.nSamples);
else
Data{i} = read_bdf(datafilename,hdr,1,hdr.nSamples);
end
end
EEG.times = [1:EEG.pnts]*1000/EEG.srate;
for j=1:size(T0,1)
startsecs(j) = T0(j,1)*946080000+T0(j,2)*2592000+T0(j,3)*86400+T0(j,4)*3600+T0(j,5)*60+T0(j,6);
end
[startsecs,indexsorted] = sort(startsecs);
T0 = T0(indexsorted,:);
if datafilelength > 1
for k = 1:datafilelength
EEG.data = [EEG.data,Data{indexsorted(k)}];
end
end
etc.T0 = T0(1,:);
EEG.etc = etc;
%read event
ind = find(ismember(filename,'evt.bdf'));
if numel(ind)
eventfilename = [pathname filename{ind}];
hdr = read_bdf(eventfilename);
evt = hdr.event;
evt = cell2mat(evt);
EEG.event = struct([]);
if isstruct(evt)
for i = 1:size(evt,2)
event = struct('type',evt(i).eventvalue,'latency',evt(i).offset_in_sec*EEG.srate);
EEG.event = [EEG.event;event];
end
end
%shift event latency for multiple data files only
if datafilelength > 1
startpnts = (startsecs-startsecs(1))*EEG.srate; %relative time
endpnts = zeros(1,datafilelength);
pausepnts = 0;
Event = struct([]);
datapnts = datapnts(indexsorted);
for j = 1:datafilelength
endpnts(j) = startpnts(j)+datapnts(j);
for k = 1:length(EEG.event)
if startpnts(j) <= EEG.event(k).latency && endpnts(j) >= EEG.event(k).latency
if j > 1
pausepnts = pausepnts+(startpnts(j)-endpnts(j-1));
EEG.event(k).latency = EEG.event(k).latency-pausepnts;
end
Event = [Event;EEG.event(k)];
end
end
if j < datafilelength
eventBoundary = struct('type','boundary','latency',endpnts(j)+1);
Event = [Event;eventBoundary];
end
end
EEG.event = Event;
end
elseif exist([pathname 'evt.bdf'],'file') == 0 && hdr.Annotation == 0
EEG.event = struct([]);
end
%read mems data and combine with EEG data
memstype = {'acc.edf','gyro.edf','mag.edf'};
for gg = 1:length(memstype)
%initialize
indexmems = [];
T0mems = [];
memsData = {};
mems.data = [];
%find the index of the mems file
for ii = 1:length(filename)
lennametype = length(memstype{gg})-4; %ignore '.edf'
if length(filename{ii}) >= lennametype
if isequal(filename{ii}(1:lennametype), memstype{gg}(1:lennametype))
indexmems = [indexmems, ii];
end
end
end
filelength = length(indexmems);
if filelength >0
for ii = 1:filelength
memsfilename = [pathname filename{indexmems(ii)}];
%read header
hdrmems = read_bdf(memsfilename);
chanlocs= hdrmems.chanlocs;
nbchan = hdrmems.nChans;
T0mems = [T0mems; hdrmems.T0];
%read data
data = read_bdf(memsfilename,hdrmems,1,hdrmems.nSamples);
%resample
datarsp = [];
for jj = 1:nbchan
datamems = resample(data(jj,:), EEG.srate, hdrmems.Fs);
datarsp = [datarsp;datamems];
end
if filelength == 1
mems.data = datarsp;
else
memsData{ii} = datarsp;
end
end
if filelength > 1
%sort the mems data files based on start time (T0)
for aa=1:size(T0mems,1)
startsecs(aa) = T0mems(aa,1)*946080000+T0mems(aa,2)*2592000+T0mems(aa,3)*86400+T0mems(aa,4)*3600+T0mems(aa,5)*60+T0mems(aa,6);
end
[startsecs,indexsorted] = sort(startsecs);
for bb = 1:filelength
mems.data = [mems.data,memsData{indexsorted(bb)}];
end
end
%make sure EEG and mems data have same length
nbpnts = size(mems.data,2) ;
if nbpnts == EEG.pnts
EEG.data = [EEG.data; mems.data];
elseif nbpnts > EEG.pnts
EEG.data = [EEG.data; mems.data(:,1:EEG.pnts)];
elseif nbpnts < EEG.pnts
EEG.data = [EEG.data(:,1:hdr.nSamples); mems.data];
end
EEG.chanlocs = [EEG.chanlocs ; chanlocs ];
EEG.nbchan = EEG.nbchan + nbchan;
end
end
else
error('Please select EEG datasets with the filenames starting by "data" ')
end
function dat = read_bdf(filename, hdr, begsample, endsample, chanindx)
if nargin==1
% read the header, this code is from EEGLAB's openbdf
FILENAME = filename;
% defines Seperator for Subdirectories
SLASH='/';
BSLASH=char(92);
cname=computer;
if cname(1:2)=='PC' SLASH=BSLASH; end;
fid=fopen(FILENAME,'r','ieee-le');
if fid<0
fprintf(2,['Error LOADEDF: File ' FILENAME ' not found\n']);
return;
end;
EDF.FILE.FID=fid;
EDF.FILE.OPEN = 1;
EDF.FileName = FILENAME;
PPos=min([max(find(FILENAME=='.')) length(FILENAME)+1]);
SPos=max([0 find((FILENAME=='/') | (FILENAME==BSLASH))]);
EDF.FILE.Ext = FILENAME(PPos+1:length(FILENAME));
EDF.FILE.Name = FILENAME(SPos+1:PPos-1);
if SPos==0
EDF.FILE.Path = pwd;
else
EDF.FILE.Path = FILENAME(1:SPos-1);
end;
EDF.FileName = [EDF.FILE.Path SLASH EDF.FILE.Name '.' EDF.FILE.Ext];
H1=char(fread(EDF.FILE.FID,256,'char')'); %
EDF.FILETYPE = H1(1); % 1 Byte bdf or edf
hdr.filetype = str2num(EDF.FILETYPE);
EDF.VERSION=H1(1:8); % 8 Byte Versionsnummer
%if 0 fprintf(2,'LOADEDF: WARNING Version EDF Format %i',ver); end;
EDF.PID = deblank(H1(9:88)); % 80 Byte local patient identification
EDF.RID = deblank(H1(89:168)); % 80 Byte local recording identification
%EDF.H.StartDate = H1(169:176); % 8 Byte
%EDF.H.StartTime = H1(177:184); % 8 Byte
EDF.T0=[str2num(H1(168+[7 8])) str2num(H1(168+[4 5])) str2num(H1(168+[1 2])) str2num(H1(168+[9 10])) str2num(H1(168+[12 13])) str2num(H1(168+[15 16])) ];
hdr.T0 = EDF.T0;
% Y2K compatibility until year 2090
if EDF.VERSION(1)=='0'
if EDF.T0(1) < 91
EDF.T0(1)=2000+EDF.T0(1);
else
EDF.T0(1)=1900+EDF.T0(1);
end;
else ;
% in a future version, this is hopefully not needed
end;
EDF.HeadLen = str2num(H1(185:192)); % 8 Byte Length of Header
% reserved = H1(193:236); % 44 Byte
EDF.NRec = str2num(H1(237:244)); % 8 Byte # of data records
EDF.Dur = str2num(H1(245:252)); % 8 Byte # duration of data record in sec
EDF.NS = str2num(H1(253:256)); % 8 Byte # of channels
EDF.Label = char(fread(EDF.FILE.FID,[16,EDF.NS],'char')'); %labels of the channels
EDF.Transducer = char(fread(EDF.FILE.FID,[80,EDF.NS],'char')');
EDF.PhysDim = char(fread(EDF.FILE.FID,[8,EDF.NS],'char')');
EDF.PhysMin= str2num(char(fread(EDF.FILE.FID,[8,EDF.NS],'char')'));
EDF.PhysMax= str2num(char(fread(EDF.FILE.FID,[8,EDF.NS],'char')'));
EDF.DigMin = str2num(char(fread(EDF.FILE.FID,[8,EDF.NS],'char')'));
EDF.DigMax = str2num(char(fread(EDF.FILE.FID,[8,EDF.NS],'char')'));
% check validity of DigMin and DigMax
if (length(EDF.DigMin) ~= EDF.NS)
fprintf(2,'Warning OPENEDF: Failing Digital Minimum\n');
EDF.DigMin = -(2^15)*ones(EDF.NS,1);
end
if (length(EDF.DigMax) ~= EDF.NS)
fprintf(2,'Warning OPENEDF: Failing Digital Maximum\n');
EDF.DigMax = (2^15-1)*ones(EDF.NS,1);
end
if (any(EDF.DigMin >= EDF.DigMax))
fprintf(2,'Warning OPENEDF: Digital Minimum larger than Maximum\n');
end
% check validity of PhysMin and PhysMax
if (length(EDF.PhysMin) ~= EDF.NS)
fprintf(2,'Warning OPENEDF: Failing Physical Minimum\n');
EDF.PhysMin = EDF.DigMin;
end
if (length(EDF.PhysMax) ~= EDF.NS)
fprintf(2,'Warning OPENEDF: Failing Physical Maximum\n');
EDF.PhysMax = EDF.DigMax;
end
if (any(EDF.PhysMin >= EDF.PhysMax))
fprintf(2,'Warning OPENEDF: Physical Minimum larger than Maximum\n');
EDF.PhysMin = EDF.DigMin;
EDF.PhysMax = EDF.DigMax;
end
EDF.PreFilt= char(fread(EDF.FILE.FID,[80,EDF.NS],'char')'); %
tmp = fread(EDF.FILE.FID,[8,EDF.NS],'char')'; % samples per data record
EDF.SPR = str2num(char(tmp)); % samples per data record
fseek(EDF.FILE.FID,32*EDF.NS,0);
EDF.Cal = (EDF.PhysMax-EDF.PhysMin)./(EDF.DigMax-EDF.DigMin);
EDF.Off = EDF.PhysMin - EDF.Cal .* EDF.DigMin;
tmp = find(EDF.Cal < 0);
EDF.Cal(tmp) = ones(size(tmp));
EDF.Off(tmp) = zeros(size(tmp));
EDF.Calib=[EDF.Off';(diag(EDF.Cal))];
%EDF.Calib=sparse(diag([1; EDF.Cal]));
%EDF.Calib(1,2:EDF.NS+1)=EDF.Off';
EDF.SampleRate = EDF.SPR / EDF.Dur;
EDF.FILE.POS = ftell(EDF.FILE.FID);
if EDF.NRec == -1 % unknown record size, determine correct NRec
fseek(EDF.FILE.FID, 0, 'eof');
endpos = ftell(EDF.FILE.FID);
EDF.NRec = floor((endpos - EDF.FILE.POS) / (sum(EDF.SPR) * 2));
fseek(EDF.FILE.FID, EDF.FILE.POS, 'bof');
H1(237:244)=sprintf('%-8i',EDF.NRec); % write number of records
end;
EDF.Chan_Select=(EDF.SPR==max(EDF.SPR));
for k=1:EDF.NS
if EDF.Chan_Select(k)
EDF.ChanTyp(k)='N';
else
EDF.ChanTyp(k)=' ';
end;
if findstr(upper(EDF.Label(k,:)),'ECG')
EDF.ChanTyp(k)='C';
elseif findstr(upper(EDF.Label(k,:)),'EKG')
EDF.ChanTyp(k)='C';
elseif findstr(upper(EDF.Label(k,:)),'EEG')
EDF.ChanTyp(k)='E';
elseif findstr(upper(EDF.Label(k,:)),'EOG')
EDF.ChanTyp(k)='O';
elseif findstr(upper(EDF.Label(k,:)),'EMG')
EDF.ChanTyp(k)='M';
end;
end;
EDF.AS.spb = sum(EDF.SPR); % Samples per Block
hdr.Annotation = 0;
hdr.AnnotationChn = -1;
if any(EDF.SampleRate~=EDF.SampleRate(1))
chn_indx = find(EDF.SampleRate~=EDF.SampleRate(1));
if(length(chn_indx) > 0 && (strcmp(EDF.Label(chn_indx,:),repmat('BDF Annotations',length(chn_indx),1))) || strcmp(EDF.Label(chn_indx,:),repmat('BDF Annotations ',length(chn_indx),1)))
disp('BDF+ file format detected, the BDF Annotation channel will be skipped when reading data');
hdr.Annotation = 1;
hdr.AnnotationChn = chn_indx;
EDF.SampleRateOrg = EDF.SampleRate;
EDF.SampleRate(chn_indx) = EDF.SampleRate(1);%force it to be the same as others, in order to pass the FieldTrip test
EDF.Label(chn_indx,:) = [];%delete the info about the Annotation channel
EDF.NS = EDF.NS - length(chn_indx);
else
error('Channels with different sampling rate but not BDF Annotation found');
end
end
%read all the events as well
if(hdr.Annotation)
epoch_total = EDF.Dur * sum(EDF.SampleRateOrg); %the number of total data per record, usually one second
% allocate memory to hold the data
n_annotation = sum(EDF.Dur*EDF.SampleRateOrg(hdr.AnnotationChn)); %the number of annotation per record
dat_event = zeros(EDF.NRec,n_annotation*3);
% read and concatenate all required data epochs
for i=1:EDF.NRec
%offset: the bit index right before the current first event
offset = EDF.HeadLen + (i-1)*epoch_total*3 + EDF.Dur*sum(EDF.SampleRateOrg(1:EDF.NS))*3;
%extract the bits for events
buf = readEvents(filename, offset, n_annotation); % see below in subfunction
% buf = readLowLevel(filename, offset, n_annotation);
dat_event(i,:) = buf;
end
%decoding the events
event_cnt = 1;
event = [];
for i = 1:EDF.NRec %last index
char20_index = find(dat_event(i,:)==20);
TAL_start = char20_index(2) + 1;%the first event right after the second char 20
char20_index = char20_index(3:end);%remove the first two 20 (belonging to the TAL time index)
num_event = length(char20_index)/2;%two char 20 per event
if(num_event == 0)
continue;
end
for j = 1:num_event
%structure of a single event: [offset in sec] [char21][Duration in sec] [char20][Trigger Code] [char20][char0]
%the field [char21][Duration in sec] can be skipped
%multiple events can be stored within one Annotation block, the last ends with [char0][char0]
singleTAL = dat_event(i,TAL_start:char20_index(2*j)-1);
char21_one = find(singleTAL == 21);
char20_one = find(singleTAL == 20);
event{event_cnt}.eventtype = 'trigger';%fixed info
event{event_cnt}.eventvalue = char(singleTAL(char20_one+1:end));%trigger code, char
if(~isempty(char21_one))%if Duration field exist
event{event_cnt}.offset_in_sec = str2num(char(singleTAL(1:char21_one-1)));%in sec
event{event_cnt}.duration = str2num(char(singleTAL(char21_one+1:char20_one-1)));%in sec;
else%if no Duration field
event{event_cnt}.offset_in_sec = str2num(char(singleTAL(1:char20_one-1)));%in sec
event{event_cnt}.duration = 0;
end
event{event_cnt}.offset = round(event{event_cnt}.offset_in_sec*EDF.SampleRateOrg(1));%in sampling points
event_cnt = event_cnt + 1;
TAL_start = char20_index(2*j) + 1;
if(dat_event(i,char20_index(2*j)+1) ~= 0)%check if the TAL is complete
error('BDF+ event error');
end
end
%check if all TALs are read
if ~(dat_event(i,char20_index(2*num_event)+1) == 0 && dat_event(i,char20_index(2*num_event)+2) == 0)
error('BDF+ final event error');
end
end
hdr.event = event;
end
% close the file
fclose(EDF.FILE.FID);
% convert the header to Fieldtrip-style
hdr.Fs = EDF.SampleRate(1);
hdr.nChans = EDF.NS;
hdr.label = cellstr(EDF.Label);
hdr.chanlocs = struct([]);
for i = 1:hdr.nChans
chanlocs = struct('labels',hdr.label(i),'ref',[],'theta',[],'radius',[],'X',[],'Y',[],'Z',[],'sph_theta',[],'sph_phi',[],'sph_radius',[],'type',[],'urchan',[] );
hdr.chanlocs = [hdr.chanlocs;chanlocs];
end
% it is continuous data, therefore append all records in one trial
hdr.nTrials = 1;
hdr.nSamples = EDF.NRec * EDF.Dur * EDF.SampleRate(1);
hdr.nSamplesPre = 0;
hdr.orig = EDF;
% return the header
dat = hdr;
else
% read the data
% retrieve the original header
EDF = hdr.orig;
% determine the trial containing the begin and end sample
epochlength = EDF.Dur * EDF.SampleRate(1);
begepoch = floor((begsample-1)/epochlength) + 1;
endepoch = floor((endsample-1)/epochlength) + 1;
nepochs = endepoch - begepoch + 1;
nchans = EDF.NS;
if(hdr.Annotation)
epoch_total = sum(EDF.Dur * EDF.SampleRateOrg);
else
epoch_total = EDF.Dur * EDF.SampleRate(1)*nchans;
end
epoch_data = EDF.Dur * EDF.SampleRate(1)*nchans;
%TODO HERE
if nargin<5
chanindx = 1:nchans;
end
if(hdr.Annotation)
for tmp_indx = 1:length(chanindx)
%make this revision as the Annotation channel is invisible to the
%users, hereby channel index (by user) larger than the Annotation
%channel should increase their index by 1
if(chanindx(tmp_indx) >= hdr.AnnotationChn)
chanindx(tmp_indx) = chanindx(tmp_indx) + 1;
end
end
end
% allocate memory to hold the data
dat = zeros(length(chanindx),nepochs*epochlength);
% read and concatenate all required data epochs
for i=begepoch:endepoch%number of records
if hdr.filetype == 0
offset = EDF.HeadLen + (i-1)*epoch_total*2;
else
offset = EDF.HeadLen + (i-1)*epoch_total*3;
end
% read the data from all channels and then select the desired channels
buf = readLowLevel(filename, offset, epoch_data,hdr.filetype); % see below in subfunction
buf = reshape(buf, epochlength, nchans);
dat(:,((i-begepoch)*epochlength+1):((i-begepoch+1)*epochlength)) = buf(:,chanindx)';
end
% select the desired samples
begsample = begsample - (begepoch-1)*epochlength; % correct for the number of bytes that were skipped
endsample = endsample - (begepoch-1)*epochlength; % correct for the number of bytes that were skipped
dat = dat(:, begsample:endsample);
% Calibrate the data
calib = diag(EDF.Cal(chanindx));
if length(chanindx)>1
% using a sparse matrix speeds up the multiplication
dat = sparse(calib) * dat;
else
% in case of one channel the sparse multiplication would result in a sparse array
dat = calib * dat;
end
end
% SUBFUNCTION for reading the 24 bit values
function buf = readLowLevel(filename, offset, numwords,filetype)
% if offset < 2*1024^3
% % use the external mex file, only works for <2GB
% buf = read_24bit(filename, offset, numwords);
% % this would be the only difference between the bdf and edf implementation
% % buf = read_16bit(filename, offset, numwords);
% else
% use plain matlab, thanks to Philip van der Broek
fp = fopen(filename,'r','ieee-le');
status = fseek(fp, offset, 'bof');
if status
error(['failed seeking ' filename]);
end
if filetype == 0
[buf,num] = fread(fp,numwords,'bit16=>double'); %edf
else
[buf,num] = fread(fp,numwords,'bit24=>double'); %bdf
end
fclose(fp);
if (num<numwords)
error(['failed opening ' filename]);
return
% end
end
function buf = readEvents(filename, offset, numwords)
% use plain matlab, thanks to Philip van der Broek
fp = fopen(filename,'r','ieee-le');
status = fseek(fp, offset, 'bof');
if status
error(['failed seeking ' filename]);
end
[buf,num] = fread(fp,numwords*3,'uint8=>char');
fclose(fp);
if (num<numwords)
error(['failed opening ' filename]);
return
end
data = readbdf(‘filename.bdf’);
其中filename.bdf是待转换的BDF文件的名称,包括文件扩展名。函数执行后,会将BDF文件中的数据读取到变量data中。
- 查看和处理数据
查看data变量的结构,了解其包含的字段和数据内容。通常,data会包含信号数据矩阵、采样率、通道名称等信息。例如:
disp(data);
根据需要对data中的数据进行进一步处理,如滤波、去噪、特征提取等操作。
- 保存为MAT文件
使用MATLAB的save函数将处理后的数据保存为MAT文件:
matlab复制
save(‘output.mat’, ‘data’);
这样,就成功将BDF文件转换并保存为MAT文件,文件名为output.mat,其中包含了变量data及其对应的数据。
(三)注意事项
确保readbdf函数与你的MATLAB版本兼容,否则可能出现调用错误。
在调用函数前,检查BDF文件的完整性和格式是否正确,避免因文件损坏导致读取失败。
如果BDF文件中包含特殊格式的数据或注释信息,需提前了解readbdf函数对这些内容的处理方式,必要时可对函数进行适当修改以满足特定需求。
三、使用EEGLAB界面操作进行转换
(一)EEGLAB简介
EEGLAB是一个基于MATLAB的开源工具箱,专门用于脑电图(EEG)等生物医学信号的分析。它提供了丰富的图形用户界面(GUI)操作功能,包括数据导入、预处理、分析和可视化等,其中也支持BDF文件的导入和转换为MAT文件的操作。
(二)操作步骤
- 安装EEGLAB
如果尚未安装EEGLAB,可以通过MATLAB的Add-On Explorer或EEGLAB官方网站下载并安装版本V13.6.5。安装完成后,在MATLAB命令窗口输入eeglab启动EEGLAB界面。
此处点击下载:EEGlabv13.6.5
- 导入BDF文件
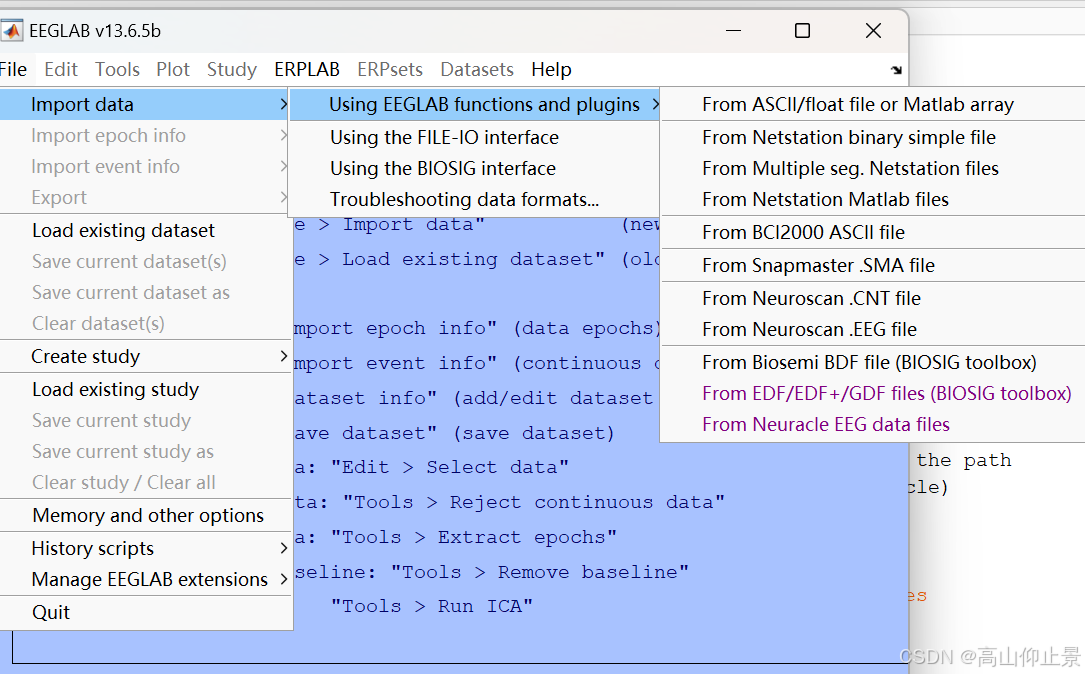
在EEGLAB主界面中,点击“File”菜单,选择“Import data”选项,然后在弹出的子菜单中选择“Import from a file”。
在文件选择对话框中,浏览并找到待转换的BDF文件,选中后点击“Open”按钮。EEGLAB会自动读取BDF文件中的数据,并将其加载到EEGLAB的数据结构中。
- 查看和编辑数据信息
导入数据后,可以在EEGLAB的“Dataset information”窗口中查看数据的基本信息,如采样率、通道数、时间长度等。如果需要,还可以对数据进行编辑和预处理操作,如重命名通道、设置参考电极等。
读取到的数据最终在变量工作区域,找到EEG(struct)这个结构体变量。
- 保存为MAT文件
完成数据查看和编辑后,点击“File”菜单,选择“Save current dataset”选项。在弹出的保存对话框中,选择保存路径和文件名,文件扩展名默认为.mat,这是EEGLAB的数据文件格式。
(三)注意事项
在导入BDF文件时,如果文件中包含特殊格式的数据或异常值,可能导致导入失败或数据错误。此时,可尝试对BDF文件进行预处理或检查文件格式是否符合EEGLAB的要求。
EEGLAB界面操作相对直观,但在进行复杂的数据处理和分析时,可能不如编写MATLAB脚本灵活。对于需要进行大量重复操作或自定义分析的情况,建议结合使用readbdf函数和MATLAB编程。
保存为.set文件后,如果需要在其他不安装EEGLAB的MATLAB环境中使用数据,可能需要对数据进行进一步处理和转换,以提取出所需的信号数据矩阵等关键信息。
四、总结
本文介绍了两种将BDF文件转换为MAT文件的方法:使用自定义函数readbdf和利用EEGLAB界面操作。readbdf函数适用于熟悉MATLAB编程的用户,能够灵活地对数据进行处理和分析;而EEGLAB界面操作则更加直观方便,适合初学者或需要进行简单数据处理的情况。在实际应用中,可根据具体需求和个人习惯选择合适的方法进行BDF到MAT的转换,以便更好地开展生物医学信号分析工作。