目录
时空图神经网络(STGCN)结构
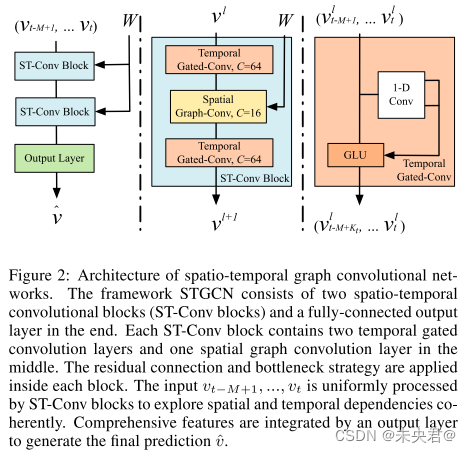
时空图神经网络(STGCN)能够捕获输入图结构数据中的时空关联性特征,STGCN由时空卷积模块(spatio-temporal convolutional blocks)组成,该模块在时域上使用门控卷积(gated convolution layer)提取数据的时间相关性,在空间域上应用图卷积(graph convolution layer)提取数据的内在联系。时空图神经网络(STGCN)的示意图如下:
图卷积和门控卷积原理不再多说,具体可以参考其他博客,直接上代码。
时空图神经网络(STGCN)代码
stgcn.py
import math
import torch
import torch.nn as nn
import torch.nn.functional as F
class TimeBlock(nn.Module):
"""
Neural network block that applies a temporal convolution to each node of
a graph in isolation.
"""
def __init__(self, in_channels, out_channels, kernel_size=3):
"""
:param in_channels: Number of input features at each node in each time
step.
:param out_channels: Desired number of output channels at each node in
each time step.
:param kernel_size: Size of the 1D temporal kernel.
"""
super(TimeBlock, self).__init__()
self.conv1 = nn.Conv2d(in_channels, out_channels, (1, kernel_size))
self.conv2 = nn.Conv2d(in_channels, out_channels, (1, kernel_size))
self.conv3 = nn.Conv2d(in_channels, out_channels, (1, kernel_size))
def forward(self, X):
"""
:param X: Input data of shape (batch_size, num_nodes, num_timesteps,
num_features=in_channels)
:return: Output data of shape (batch_size, num_nodes,
num_timesteps_out, num_features_out=out_channels)
"""
# Convert into NCHW format for pytorch to perform convolutions.
X = X.permute(0, 3, 1, 2)
temp = self.conv1(X) + torch.sigmoid(self.conv2(X))
out = F.relu(temp + self.conv3(X))
# Convert back from NCHW to NHWC
out = out.permute(0, 2, 3, 1)
return out
class STGCNBlock(nn.Module):
"""
Neural network block that applies a temporal convolution on each node in
isolation, followed by a graph convolution, followed by another temporal
convolution on each node.
"""
def __init__(self, in_channels, spatial_channels, out_channels,
num_nodes):
"""
:param in_channels: Number of input features at each node in each time
step.
:param spatial_channels: Number of output channels of the graph
convolutional, spatial sub-block.
:param out_channels: Desired number of output features at each node in
each time step.
:param num_nodes: Number of nodes in the graph.
"""
super(STGCNBlock, self).__init__()
self.temporal1 = TimeBlock(in_channels=in_channels,
out_channels=out_channels)
self.Theta1 = nn.Parameter(torch.FloatTensor(out_channels,
spatial_channels))
self.temporal2 = TimeBlock(in_channels=spatial_channels,
out_channels=out_channels)
self.batch_norm = nn.BatchNorm2d(num_nodes)
self.reset_parameters()
def reset_parameters(self):
stdv = 1. / math.sqrt(self.Theta1.shape[1])
self.Theta1.data.uniform_(-stdv, stdv)
def forward(self, X, A_hat):
"""
:param X: Input data of shape (batch_size, num_nodes, num_timesteps,
num_features=in_channels).
:param A_hat: Normalized adjacency matrix.
:return: Output data of shape (batch_size, num_nodes,
num_timesteps_out, num_features=out_channels).
"""
t = self.temporal1(X)
lfs = torch.einsum("ij,jklm->kilm", [A_hat, t.permute(1, 0, 2, 3)])
# t2 = F.relu(torch.einsum("ijkl,lp->ijkp", [lfs, self.Theta1]))
t2 = F.relu(torch.matmul(lfs, self.Theta1))
t3 = self.temporal2(t2)
return self.batch_norm(t3)
# return t3
class STGCN(nn.Module):
"""
Spatio-temporal graph convolutional network as described in
https://arxiv.org/abs/1709.04875v3 by Yu et al.
Input should have shape (batch_size, num_nodes, num_input_time_steps,
num_features).
"""
def __init__(self, num_nodes, num_features, num_timesteps_input,
num_timesteps_output):
"""
:param num_nodes: Number of nodes in the graph.
:param num_features: Number of features at each node in each time step.
:param num_timesteps_input: Number of past time steps fed into the
network.
:param num_timesteps_output: Desired number of future time steps
output by the network.
"""
super(STGCN, self).__init__()
self.block1 = STGCNBlock(in_channels=num_features, out_channels=64,
spatial_channels=16, num_nodes=num_nodes)
self.block2 = STGCNBlock(in_channels=64, out_channels=64,
spatial_channels=16, num_nodes=num_nodes)
self.last_temporal = TimeBlock(in_channels=64, out_channels=64)
self.fully = nn.Linear((num_timesteps_input - 2 * 5) * 64,
num_timesteps_output)
def forward(self, A_hat, X):
"""
:param X: Input data of shape (batch_size, num_nodes, num_timesteps,
num_features=in_channels).
:param A_hat: Normalized adjacency matrix.
"""
out1 = self.block1(X, A_hat)
out2 = self.block2(out1, A_hat)
out3 = self.last_temporal(out2)
out4 = self.fully(out3.reshape((out3.shape[0], out3.shape[1], -1)))
return
main.py
import os
import argparse
import pickle as pk
import numpy as np
import matplotlib.pyplot as plt
import torch
import torch.nn as nn
from stgcn import STGCN
from utils import generate_dataset, load_metr_la_data, get_normalized_adj
use_gpu = False
num_timesteps_input = 12
num_timesteps_output = 3
epochs = 1000
batch_size = 50
parser = argparse.ArgumentParser(description='STGCN')
parser.add_argument('--enable-cuda', action='store_true',
help='Enable CUDA')
args = parser.parse_args()
args.device = None
if args.enable_cuda and torch.cuda.is_available():
args.device = torch.device('cuda')
else:
args.device = torch.device('cpu')
def train_epoch(training_input, training_target, batch_size):
"""
Trains one epoch with the given data.
:param training_input: Training inputs of shape (num_samples, num_nodes,
num_timesteps_train, num_features).
:param training_target: Training targets of shape (num_samples, num_nodes,
num_timesteps_predict).
:param batch_size: Batch size to use during training.
:return: Average loss for this epoch.
"""
permutation = torch.randperm(training_input.shape[0])
epoch_training_losses = []
for i in range(0, training_input.shape[0], batch_size):
net.train()
optimizer.zero_grad()
indices = permutation[i:i + batch_size]
X_batch, y_batch = training_input[indices], training_target[indices]
X_batch = X_batch.to(device=args.device)
y_batch = y_batch.to(device=args.device)
out = net(A_wave, X_batch)
loss = loss_criterion(out, y_batch)
loss.backward()
optimizer.step()
epoch_training_losses.append(loss.detach().cpu().numpy())
return sum(epoch_training_losses)/len(epoch_training_losses)
if __name__ == '__main__':
torch.manual_seed(7)
A, X, means, stds = load_metr_la_data()
split_line1 = int(X.shape[2] * 0.6)
split_line2 = int(X.shape[2] * 0.8)
train_original_data = X[:, :, :split_line1]
val_original_data = X[:, :, split_line1:split_line2]
test_original_data = X[:, :, split_line2:]
training_input, training_target = generate_dataset(train_original_data,
num_timesteps_input=num_timesteps_input,
num_timesteps_output=num_timesteps_output)
val_input, val_target = generate_dataset(val_original_data,
num_timesteps_input=num_timesteps_input,
num_timesteps_output=num_timesteps_output)
test_input, test_target = generate_dataset(test_original_data,
num_timesteps_input=num_timesteps_input,
num_timesteps_output=num_timesteps_output)
A_wave = get_normalized_adj(A)
A_wave = torch.from_numpy(A_wave)
A_wave = A_wave.to(device=args.device)
net = STGCN(A_wave.shape[0],
training_input.shape[3],
num_timesteps_input,
num_timesteps_output).to(device=args.device)
optimizer = torch.optim.Adam(net.parameters(), lr=1e-3)
loss_criterion = nn.MSELoss()
training_losses = []
validation_losses = []
validation_maes = []
for epoch in range(epochs):
loss = train_epoch(training_input, training_target,
batch_size=batch_size)
training_losses.append(loss)
# Run validation
with torch.no_grad():
net.eval()
val_input = val_input.to(device=args.device)
val_target = val_target.to(device=args.device)
out = net(A_wave, val_input)
val_loss = loss_criterion(out, val_target).to(device="cpu")
validation_losses.append(np.asscalar(val_loss.detach().numpy()))
out_unnormalized = out.detach().cpu().numpy()*stds[0]+means[0]
target_unnormalized = val_target.detach().cpu().numpy()*stds[0]+means[0]
mae = np.mean(np.absolute(out_unnormalized - target_unnormalized))
validation_maes.append(mae)
out = None
val_input = val_input.to(device="cpu")
val_target = val_target.to(device="cpu")
print("Training loss: {}".format(training_losses[-1]))
print("Validation loss: {}".format(validation_losses[-1]))
print("Validation MAE: {}".format(validation_maes[-1]))
plt.plot(training_losses, label="training loss")
plt.plot(validation_losses, label="validation loss")
plt.legend()
plt.show()
checkpoint_path = "checkpoints/"
if not os.path.exists(checkpoint_path):
os.makedirs(checkpoint_path)
with open("checkpoints/losses.pk", "wb") as fd:
pk.dump((training_losses, validation_losses, validation_maes), fd)
utils.py
import os
import zipfile
import numpy as np
import torch
def load_metr_la_data():
if (not os.path.isfile("data/adj_mat.npy")
or not os.path.isfile("data/node_values.npy")):
with zipfile.ZipFile("data/METR-LA.zip", 'r') as zip_ref:
zip_ref.extractall("data/")
A = np.load("data/adj_mat.npy")
X = np.load("data/node_values.npy").transpose((1, 2, 0))
X = X.astype(np.float32)
# Normalization using Z-score method
means = np.mean(X, axis=(0, 2))
X = X - means.reshape(1, -1, 1)
stds = np.std(X, axis=(0, 2))
X = X / stds.reshape(1, -1, 1)
return A, X, means, stds
def get_normalized_adj(A):
"""
Returns the degree normalized adjacency matrix.
"""
A = A + np.diag(np.ones(A.shape[0], dtype=np.float32))
D = np.array(np.sum(A, axis=1)).reshape((-1,))
D[D <= 10e-5] = 10e-5 # Prevent infs
diag = np.reciprocal(np.sqrt(D))
A_wave = np.multiply(np.multiply(diag.reshape((-1, 1)), A),
diag.reshape((1, -1)))
return A_wave
def generate_dataset(X, num_timesteps_input, num_timesteps_output):
"""
Takes node features for the graph and divides them into multiple samples
along the time-axis by sliding a window of size (num_timesteps_input+
num_timesteps_output) across it in steps of 1.
:param X: Node features of shape (num_vertices, num_features,
num_timesteps)
:return:
- Node features divided into multiple samples. Shape is
(num_samples, num_vertices, num_features, num_timesteps_input).
- Node targets for the samples. Shape is
(num_samples, num_vertices, num_features, num_timesteps_output).
"""
# Generate the beginning index and the ending index of a sample, which
# contains (num_points_for_training + num_points_for_predicting) points
indices = [(i, i + (num_timesteps_input + num_timesteps_output)) for i
in range(X.shape[2] - (
num_timesteps_input + num_timesteps_output) + 1)]
# Save samples
features, target = [], []
for i, j in indices:
features.append(
X[:, :, i: i + num_timesteps_input].transpose(
(0, 2, 1)))
target.append(X[:, 0, i + num_timesteps_input: j])
return torch.from_numpy(np.array(features)), \
torch.from_numpy(np.array(target))