该方法基于python进行时间序列的小波分析并出图(包括功率谱图和小波分解后的图)。
默认的小波为morlet小波。
该代码由 Evgeniya Predybaylo 博士提供:
https://github.com/chris-torrence/wavelets
https://atoc.colorado.edu/research/wavelets/
Copyright © 1995-2004, Christopher Torrence and Gilbert P.Compo
Python version of the code is written by Evgeniya Predybaylo in 2014
代码及示例数据:(打包后的文件,具体代码贴在文章最后)
https://download.csdn.net/download/qq_32832803/13944688
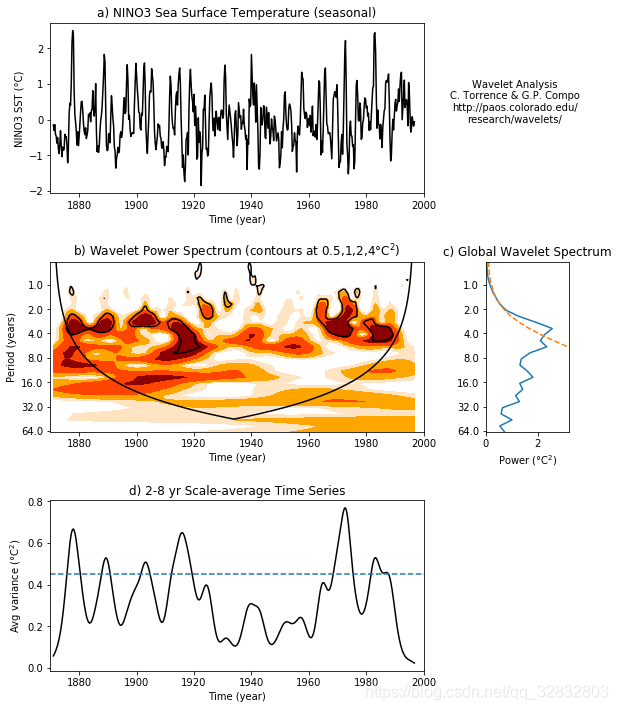
出图效果:
- 图a表示源数据的时间序列。
- 图b和图c表示功率谱图和全部时间长度的功率谱分布。图b粗黑线包围的范围表示通过了0.05显著性检验;U型细黑线为影响锥曲线(COI),在该曲线以外的功率谱由于受到边界效应的影响而不予考虑。图c分析时舍弃红色线以外的部分,红色线以内波峰高处为主要的周期。
- 图d表示2-8年的窗口平均。
频率谱图的分析参考:
叶许春, 许崇育, 张丹, 李相虎. 长江中下游夏季降水变化与亚洲夏季风系统的关系[J]. 地理科学, 2018, 38(7): 1174-1182 https://doi.org/10.13249/j.cnki.sgs.2018.07.019
waveletAnalysis.py
import numpy as np
from waveletFunctions import wavelet, wave_signif
import matplotlib.pylab as plt
from matplotlib.gridspec import GridSpec
import matplotlib.ticker as ticker
from mpl_toolkits.axes_grid1 import make_axes_locatable
__author__ = 'Evgeniya Predybaylo'
# WAVETEST Example Python script for WAVELET, using NINO3 SST dataset
#
# See "http://paos.colorado.edu/research/wavelets/"
# The Matlab code written January 1998 by C. Torrence is modified to Python by Evgeniya Predybaylo, December 2014
#
# Modified Oct 1999, changed Global Wavelet Spectrum (GWS) to be sideways,
# changed all "log" to "log2", changed logarithmic axis on GWS to
# a normal axis.
# ------------------------------------------------------------------------------------------------------------------
# READ THE DATA
sst = np.loadtxt('sst_nino3.dat') # input SST time series
sst = sst - np.mean(sst)
variance = np.std(sst, ddof=1) ** 2
print("variance = ", variance)
#----------C-O-M-P-U-T-A-T-I-O-N------S-T-A-R-T-S------H-E-R-E------------------------------------------------------
# normalize by standard deviation (not necessary, but makes it easier
# to compare with plot on Interactive Wavelet page, at
# "http://paos.colorado.edu/research/wavelets/plot/"
if 0:
variance = 1.0
sst = sst / np.std(sst, ddof=1)
n = len(sst)
dt = 0.25
time = np.arange(len(sst)) * dt + 1871.0 # construct time array
xlim = ([1870, 2000]) # plotting range
pad = 1 # pad the time series with zeroes (recommended)
dj = 0.25 # this will do 4 sub-octaves per octave
s0 = 2 * dt # this says start at a scale of 6 months
j1 = 7 / dj # this says do 7 powers-of-two with dj sub-octaves each
lag1 = 0.72 # lag-1 autocorrelation for red noise background
print("lag1 = ", lag1)
mother = 'MORLET'
# Wavelet transform:
wave, period, scale, coi = wavelet(sst, dt, pad, dj, s0, j1, mother)
power = (np.abs(wave)) ** 2 # compute wavelet power spectrum
global_ws = (np.sum(power, axis=1) / n) # time-average over all times
# Significance levels:
signif = wave_signif(([variance]), dt=dt, sigtest=0, scale=scale,
lag1=lag1, mother=mother)
sig95 = signif[:, np.newaxis].dot(np.ones(n)[np.newaxis, :]) # expand signif --> (J+1)x(N) array
sig95 = power / sig95 # where ratio > 1, power is significant
# Global wavelet spectrum & significance levels:
dof = n - scale # the -scale corrects for padding at edges
global_signif = wave_signif(variance, dt=dt, scale=scale, sigtest=1,
lag1=lag1, dof=dof, mother=mother)
# Scale-average between El Nino periods of 2--8 years
avg = np.logical_and(scale >= 2, scale < 8)
Cdelta = 0.776 # this is for the MORLET wavelet
scale_avg = scale[:, np.newaxis].dot(np.ones(n)[np.newaxis, :]) # expand scale --> (J+1)x(N) array
scale_avg = power / scale_avg # [Eqn(24)]
scale_avg = dj * dt / Cdelta * sum(scale_avg[avg, :]) # [Eqn(24)]
scaleavg_signif = wave_signif(variance, dt=dt, scale=scale, sigtest=2,
lag1=lag1, dof=([2, 7.9]), mother=mother)
#------------------------------------------------------ Plotting
#--- Plot time series
fig = plt.figure(figsize=(9, 10))
gs = GridSpec(3, 4, hspace=0.4, wspace=0.75)
plt.subplots_adjust(left=0.1, bottom=0.05, right=0.9, top=0.95, wspace=0, hspace=0)
plt.subplot(gs[0, 0:3])
plt.plot(time, sst, 'k')
plt.xlim(xlim[:])
plt.xlabel('Time (year)')
plt.ylabel('NINO3 SST (\u00B0C)')
plt.title('a) NINO3 Sea Surface Temperature (seasonal)')
plt.text(time[-1] + 35, 0.5,'Wavelet Analysis\nC. Torrence & G.P. Compo\n' +
'http://paos.colorado.edu/\nresearch/wavelets/',
horizontalalignment='center', verticalalignment='center')
#--- Contour plot wavelet power spectrum
# plt3 = plt.subplot(3, 1, 2)
plt3 = plt.subplot(gs[1, 0:3])
levels = [0, 0.5, 1, 2, 4, 999]
CS = plt.contourf(time, period, power, len(levels)) #*** or use 'contour'
im = plt.contourf(CS, levels=levels, colors=['white','bisque','orange','orangered','darkred'])
plt.xlabel('Time (year)')
plt.ylabel('Period (years)')
plt.title('b) Wavelet Power Spectrum (contours at 0.5,1,2,4\u00B0C$^2$)')
plt.xlim(xlim[:])
# 95# significance contour, levels at -99 (fake) and 1 (95# signif)
plt.contour(time, period, sig95, [-99, 1], colors='k')
# cone-of-influence, anything "below" is dubious
plt.plot(time, coi, 'k')
# format y-scale
plt3.set_yscale('log', basey=2, subsy=None)
plt.ylim([np.min(period), np.max(period)])
ax = plt.gca().yaxis
ax.set_major_formatter(ticker.ScalarFormatter())
plt3.ticklabel_format(axis='y', style='plain')
plt3.invert_yaxis()
# set up the size and location of the colorbar
# position=fig.add_axes([0.5,0.36,0.2,0.01])
# plt.colorbar(im, cax=position, orientation='horizontal') #, fraction=0.05, pad=0.5)
# plt.subplots_adjust(right=0.7, top=0.9)
#--- Plot global wavelet spectrum
plt4 = plt.subplot(gs[1, -1])
plt.plot(global_ws, period)
plt.plot(global_signif, period, '--')
plt.xlabel('Power (\u00B0C$^2$)')
plt.title('c) Global Wavelet Spectrum')
plt.xlim([0, 1.25 * np.max(global_ws)])
# format y-scale
plt4.set_yscale('log', basey=2, subsy=None)
plt.ylim([np.min(period), np.max(period)])
ax = plt.gca().yaxis
ax.set_major_formatter(ticker.ScalarFormatter())
plt4.ticklabel_format(axis='y', style='plain')
plt4.invert_yaxis()
# --- Plot 2--8 yr scale-average time series
plt.subplot(gs[2, 0:3])
plt.plot(time, scale_avg, 'k')
plt.xlim(xlim[:])
plt.xlabel('Time (year)')
plt.ylabel('Avg variance (\u00B0C$^2$)')
plt.title('d) 2-8 yr Scale-average Time Series')
plt.plot(xlim, scaleavg_signif + [0, 0], '--')
plt.show()
# end of code
waveletFunctions.py
from scipy.special._ufuncs import gammainc, gamma
import numpy as np
from scipy.optimize import fminbound
__author__ = 'Evgeniya Predybaylo'
# Copyright (C) 1995-2004, Christopher Torrence and Gilbert P.Compo
# Python version of the code is written by Evgeniya Predybaylo in 2014
#
# This software may be used, copied, or redistributed as long as it is not
# sold and this copyright notice is reproduced on each copy made. This
# routine is provided as is without any express or implied warranties
# whatsoever.
#
# Notice: Please acknowledge the use of the above software in any publications:
# Wavelet software was provided by C. Torrence and G. Compo,
# and is available at URL: http://paos.colorado.edu/research/wavelets/''.
#
# Reference: Torrence, C. and G. P. Compo, 1998: A Practical Guide to
# Wavelet Analysis. <I>Bull. Amer. Meteor. Soc.</I>, 79, 61-78.
#
# Please send a copy of such publications to either C. Torrence or G. Compo:
# Dr. Christopher Torrence Dr. Gilbert P. Compo
# Research Systems, Inc. Climate Diagnostics Center
# 4990 Pearl East Circle 325 Broadway R/CDC1
# Boulder, CO 80301, USA Boulder, CO 80305-3328, USA
# E-mail: chris[AT]rsinc[DOT]com E-mail: compo[AT]colorado[DOT]edu
#
#-------------------------------------------------------------------------------------------------------------------
# # WAVELET 1D Wavelet transform with optional significance testing
# wave, period, scale, coi = wavelet(Y, dt, pad, dj, s0, J1, mother, param)
#
# Computes the wavelet transform of the vector Y (length N),
# with sampling rate DT.
#
# By default, the Morlet wavelet (k0=6) is used.
# The wavelet basis is normalized to have total energy=1 at all scales.
#
# INPUTS:
#
# Y = the time series of length N.
# DT = amount of time between each Y value, i.e. the sampling time.
#
# OUTPUTS:
#
# WAVE is the WAVELET transform of Y. This is a complex array
# of dimensions (N,J1+1). FLOAT(WAVE) gives the WAVELET amplitude,
# ATAN(IMAGINARY(WAVE),FLOAT(WAVE) gives the WAVELET phase.
# The WAVELET power spectrum is ABS(WAVE)**2.
# Its units are sigma**2 (the time series variance).
#
# OPTIONAL INPUTS:
#
# *** Note *** if none of the optional variables is set up, then the program
# uses default values of -1.
#
# PAD = if set to 1 (default is 0), pad time series with enough zeroes to get
# N up to the next higher power of 2. This prevents wraparound
# from the end of the time series to the beginning, and also
# speeds up the FFT's used to do the wavelet transform.
# This will not eliminate all edge effects (see COI below).
#
# DJ = the spacing between discrete scales. Default is 0.25.
# A smaller # will give better scale resolution, but be slower to plot.
#
# S0 = the smallest scale of the wavelet. Default is 2*DT.
#
# J1 = the # of scales minus one. Scales range from S0 up to S0*2**(J1*DJ),
# to give a total of (J1+1) scales. Default is J1 = (LOG2(N DT/S0))/DJ.
#
# MOTHER = the mother wavelet function.
# The choices are 'MORLET', 'PAUL', or 'DOG'
#
# PARAM = the mother wavelet parameter.
# For 'MORLET' this is k0 (wavenumber), default is 6.
# For 'PAUL' this is m (order), default is 4.
# For 'DOG' this is m (m-th derivative), default is 2.
#
#
# OPTIONAL OUTPUTS:
#
# PERIOD = the vector of "Fourier" periods (in time units) that corresponds
# to the SCALEs.
#
# SCALE = the vector of scale indices, given by S0*2**(j*DJ), j=0...J1
# where J1+1 is the total # of scales.
#
# COI = if specified, then return the Cone-of-Influence, which is a vector
# of N points that contains the maximum period of useful information
# at that particular time.
# Periods greater than this are subject to edge effects.
def wavelet(Y, dt, pad=0, dj=-1, s0=-1, J1=-1, mother=-1, param=-1):
n1 = len(Y)
if s0 == -1:
s0 = 2 * dt
if dj == -1:
dj = 1. / 4.
if J1 == -1:
J1 = np.fix((np.log(n1 * dt / s0) / np.log(2)) / dj)
if mother == -1:
mother = 'MORLET'
#....construct time series to analyze, pad if necessary
x = Y - np.mean(Y)
if pad == 1:
base2 = np.fix(np.log(n1) / np.log(2) + 0.4999) # power of 2 nearest to N
x = np.concatenate((x, np.zeros((2 ** (base2 + 1) - n1).astype(np.int64))))
n = len(x)
#....construct wavenumber array used in transform [Eqn(5)]
kplus = np.arange(1, np.fix(n / 2 + 1))
kplus = (kplus * 2 * np.pi / (n * dt))
kminus = (-(kplus[0:-1])[::-1])
k = np.concatenate(([0.], kplus, kminus))
#....compute FFT of the (padded) time series
f = np.fft.fft(x) # [Eqn(3)]
#....construct SCALE array & empty PERIOD & WAVE arrays
j = np.arange(0, J1+1)
scale = s0 * 2. ** (j * dj)
wave = np.zeros(shape=(int(J1 + 1), n), dtype=complex) # define the wavelet array
# loop through all scales and compute transform
for a1 in range(0, int(J1+1)):
daughter, fourier_factor, coi, dofmin = wave_bases(mother, k, scale[a1], param)
wave[a1, :] = np.fft.ifft(f * daughter) # wavelet transform[Eqn(4)]
period = fourier_factor * scale # [Table(1)]
coi = coi * dt * np.concatenate((np.insert(np.arange(int((n1 + 1) / 2 - 1)), [0], [1E-5]),
np.insert(np.flipud(np.arange(0, n1 / 2 - 1)), [-1], [1E-5]))) # COI [Sec.3g]
wave = wave[:, :n1] # get rid of padding before returning
return wave, period, scale, coi
#-------------------------------------------------------------------------------------------------------------------
# WAVE_BASES 1D Wavelet functions Morlet, Paul, or DOG
#
# DAUGHTER,FOURIER_FACTOR,COI,DOFMIN = wave_bases(MOTHER,K,SCALE,PARAM)
#
# Computes the wavelet function as a function of Fourier frequency,
# used for the wavelet transform in Fourier space.
# (This program is called automatically by WAVELET)
#
# INPUTS:
#
# MOTHER = a string, equal to 'MORLET' or 'PAUL' or 'DOG'
# K = a vector, the Fourier frequencies at which to calculate the wavelet
# SCALE = a number, the wavelet scale
# PARAM = the nondimensional parameter for the wavelet function
#
# OUTPUTS:
#
# DAUGHTER = a vector, the wavelet function
# FOURIER_FACTOR = the ratio of Fourier period to scale
# COI = a number, the cone-of-influence size at the scale
# DOFMIN = a number, degrees of freedom for each point in the wavelet power
# (either 2 for Morlet and Paul, or 1 for the DOG)
def wave_bases(mother, k, scale, param):
n = len(k)
kplus = np.array(k > 0., dtype=float)
if mother == 'MORLET': # ----------------------------------- Morlet
if param == -1:
param = 6.
k0 = np.copy(param)
expnt = -(scale * k - k0) ** 2 / 2. * kplus
norm = np.sqrt(scale * k[1]) * (np.pi ** (-0.25)) * \
np.sqrt(n) # total energy=N [Eqn(7)]
daughter = norm * np.exp(expnt)
daughter = daughter * kplus # Heaviside step function
fourier_factor = (4 * np.pi) / (k0 + np.sqrt(2 + k0 ** 2)
) # Scale-->Fourier [Sec.3h]
coi = fourier_factor / np.sqrt(2) # Cone-of-influence [Sec.3g]
dofmin = 2 # Degrees of freedom
elif mother == 'PAUL': # -------------------------------- Paul
if param == -1:
param = 4.
m = param
expnt = -scale * k * kplus
norm = np.sqrt(scale * k[1]) * (2 ** m / np.sqrt(m *
np.prod(np.arange(1, (2 * m))))) * np.sqrt(n)
daughter = norm * ((scale * k) ** m) * np.exp(expnt) * kplus
fourier_factor = 4 * np.pi / (2 * m + 1)
coi = fourier_factor * np.sqrt(2)
dofmin = 2
elif mother == 'DOG': # -------------------------------- DOG
if param == -1:
param = 2.
m = param
expnt = -(scale * k) ** 2 / 2.0
norm = np.sqrt(scale * k[1] / gamma(m + 0.5)) * np.sqrt(n)
daughter = -norm * (1j ** m) * ((scale * k) ** m) * np.exp(expnt)
fourier_factor = 2 * np.pi * np.sqrt(2. / (2 * m + 1))
coi = fourier_factor / np.sqrt(2)
dofmin = 1
else:
print('Mother must be one of MORLET, PAUL, DOG')
return daughter, fourier_factor, coi, dofmin
#-------------------------------------------------------------------------------------------------------------------
# WAVE_SIGNIF Significance testing for the 1D Wavelet transform WAVELET
#
# SIGNIF = wave_signif(Y,DT,SCALE,SIGTEST,LAG1,SIGLVL,DOF,MOTHER,PARAM)
#
# INPUTS:
#
# Y = the time series, or, the VARIANCE of the time series.
# (If this is a single number, it is assumed to be the variance...)
# DT = amount of time between each Y value, i.e. the sampling time.
# SCALE = the vector of scale indices, from previous call to WAVELET.
#
#
# OUTPUTS:
#
# SIGNIF = significance levels as a function of SCALE
# FFT_THEOR = output theoretical red-noise spectrum as fn of PERIOD
#
#
# OPTIONAL INPUTS:
# SIGTEST = 0, 1, or 2. If omitted, then assume 0.
#
# If 0 (the default), then just do a regular chi-square test,
# i.e. Eqn (18) from Torrence & Compo.
# If 1, then do a "time-average" test, i.e. Eqn (23).
# In this case, DOF should be set to NA, the number
# of local wavelet spectra that were averaged together.
# For the Global Wavelet Spectrum, this would be NA=N,
# where N is the number of points in your time series.
# If 2, then do a "scale-average" test, i.e. Eqns (25)-(28).
# In this case, DOF should be set to a
# two-element vector [S1,S2], which gives the scale
# range that was averaged together.
# e.g. if one scale-averaged scales between 2 and 8,
# then DOF=[2,8].
#
# LAG1 = LAG 1 Autocorrelation, used for SIGNIF levels. Default is 0.0
#
# SIGLVL = significance level to use. Default is 0.95
#
# DOF = degrees-of-freedom for signif test.
# IF SIGTEST=0, then (automatically) DOF = 2 (or 1 for MOTHER='DOG')
# IF SIGTEST=1, then DOF = NA, the number of times averaged together.
# IF SIGTEST=2, then DOF = [S1,S2], the range of scales averaged.
#
# Note: IF SIGTEST=1, then DOF can be a vector (same length as SCALEs),
# in which case NA is assumed to vary with SCALE.
# This allows one to average different numbers of times
# together at different scales, or to take into account
# things like the Cone of Influence.
# See discussion following Eqn (23) in Torrence & Compo.
#
# GWS = global wavelet spectrum, a vector of the same length as scale.
# If input then this is used as the theoretical background spectrum,
# rather than white or red noise.
def wave_signif(Y, dt, scale, sigtest=0, lag1=0.0, siglvl=0.95,
dof=None, mother='MORLET', param=None, gws=None):
n1 = len(np.atleast_1d(Y))
J1 = len(scale) - 1
s0 = np.min(scale)
dj = np.log2(scale[1] / scale[0])
if n1 == 1:
variance = Y
else:
variance = np.std(Y) ** 2
# get the appropriate parameters [see Table(2)]
if mother == 'MORLET': # ---------------------------------- Morlet
empir = ([2., -1, -1, -1])
if param is None:
param = 6.
empir[1:] = ([0.776, 2.32, 0.60])
k0 = param
fourier_factor = (4 * np.pi) / (k0 + np.sqrt(2 + k0 ** 2)) # Scale-->Fourier [Sec.3h]
elif mother == 'PAUL':
empir = ([2, -1, -1, -1])
if param is None:
param = 4
empir[1:] = ([1.132, 1.17, 1.5])
m = param
fourier_factor = (4 * np.pi) / (2 * m + 1)
elif mother == 'DOG': # -------------------------------------Paul
empir = ([1., -1, -1, -1])
if param is None:
param = 2.
empir[1:] = ([3.541, 1.43, 1.4])
elif param == 6: # --------------------------------------DOG
empir[1:] = ([1.966, 1.37, 0.97])
m = param
fourier_factor = 2 * np.pi * np.sqrt(2. / (2 * m + 1))
else:
print('Mother must be one of MORLET, PAUL, DOG')
period = scale * fourier_factor
dofmin = empir[0] # Degrees of freedom with no smoothing
Cdelta = empir[1] # reconstruction factor
gamma_fac = empir[2] # time-decorrelation factor
dj0 = empir[3] # scale-decorrelation factor
freq = dt / period # normalized frequency
if gws is not None: # use global-wavelet as background spectrum
fft_theor = gws
else:
fft_theor = (1 - lag1 ** 2) / (1 - 2 * lag1 *
np.cos(freq * 2 * np.pi) + lag1 ** 2) # [Eqn(16)]
fft_theor = variance * fft_theor # include time-series variance
signif = fft_theor
if dof is None:
dof = dofmin
if sigtest == 0: # no smoothing, DOF=dofmin [Sec.4]
dof = dofmin
chisquare = chisquare_inv(siglvl, dof) / dof
signif = fft_theor * chisquare # [Eqn(18)]
elif sigtest == 1: # time-averaged significance
if len(np.atleast_1d(dof)) == 1:
dof = np.zeros(J1) + dof
dof[dof < 1] = 1
dof = dofmin * np.sqrt(1 + (dof * dt / gamma_fac / scale) ** 2) # [Eqn(23)]
dof[dof < dofmin] = dofmin # minimum DOF is dofmin
for a1 in range(0, J1 + 1):
chisquare = chisquare_inv(siglvl, dof[a1]) / dof[a1]
signif[a1] = fft_theor[a1] * chisquare
elif sigtest == 2: # time-averaged significance
if len(dof) != 2:
print('ERROR: DOF must be set to [S1,S2], the range of scale-averages')
if Cdelta == -1:
print('ERROR: Cdelta & dj0 not defined for ' +
mother + ' with param = ' + str(param))
s1 = dof[0]
s2 = dof[1]
avg = np.logical_and(scale >= 2, scale < 8) # scales between S1 & S2
navg = np.sum(np.array(np.logical_and(scale >= 2, scale < 8), dtype=int))
if navg == 0:
print('ERROR: No valid scales between ' + str(s1) + ' and ' + str(s2))
Savg = 1. / np.sum(1. / scale[avg]) # [Eqn(25)]
Smid = np.exp((np.log(s1) + np.log(s2)) / 2.) # power-of-two midpoint
dof = (dofmin * navg * Savg / Smid) * \
np.sqrt(1 + (navg * dj / dj0) ** 2) # [Eqn(28)]
fft_theor = Savg * np.sum(fft_theor[avg] / scale[avg]) # [Eqn(27)]
chisquare = chisquare_inv(siglvl, dof) / dof
signif = (dj * dt / Cdelta / Savg) * fft_theor * chisquare # [Eqn(26)]
else:
print('ERROR: sigtest must be either 0, 1, or 2')
return signif
#-------------------------------------------------------------------------------------------------------------------
# CHISQUARE_INV Inverse of chi-square cumulative distribution function (cdf).
#
# X = chisquare_inv(P,V) returns the inverse of chi-square cdf with V
# degrees of freedom at fraction P.
# This means that P*100 percent of the distribution lies between 0 and X.
#
# To check, the answer should satisfy: P==gammainc(X/2,V/2)
# Uses FMIN and CHISQUARE_SOLVE
def chisquare_inv(P, V):
if (1 - P) < 1E-4:
print('P must be < 0.9999')
if P == 0.95 and V == 2: # this is a no-brainer
X = 5.9915
return X
MINN = 0.01 # hopefully this is small enough
MAXX = 1 # actually starts at 10 (see while loop below)
X = 1
TOLERANCE = 1E-4 # this should be accurate enough
while (X + TOLERANCE) >= MAXX: # should only need to loop thru once
MAXX = MAXX * 10.
# this calculates value for X, NORMALIZED by V
X = fminbound(chisquare_solve, MINN, MAXX, args=(P, V), xtol=TOLERANCE)
MINN = MAXX
X = X * V # put back in the goofy V factor
return X # end of code
#-------------------------------------------------------------------------------------------------------------------
# CHISQUARE_SOLVE Internal function used by CHISQUARE_INV
#
# PDIFF=chisquare_solve(XGUESS,P,V) Given XGUESS, a percentile P,
# and degrees-of-freedom V, return the difference between
# calculated percentile and P.
# Uses GAMMAINC
#
# Written January 1998 by C. Torrence
# extra factor of V is necessary because X is Normalized
def chisquare_solve(XGUESS, P, V):
PGUESS = gammainc(V/2, V*XGUESS/2) # incomplete Gamma function
PDIFF = np.abs(PGUESS - P) # error in calculated P
TOL = 1E-4
if PGUESS >= 1-TOL: # if P is very close to 1 (i.e. a bad guess)
PDIFF = XGUESS # then just assign some big number like XGUESS
return PDIFF