文章目录
Unsupervised Domain Adaptation via Disentangled Representations: Application to Cross-Modality Liver Segmentation
1. Author
Junlin Yang1, Nicha C. Dvornek3, Fan Zhang2, Julius Chapiro3, MingDe Lin3, and James S. Duncan1;2;3;4
1 Department of Biomedical Engineering, Yale University, New Haven, CT, USA
2 Department of Electrical Engineering, Yale University, New Haven, CT, USA
3 Department of Radiology & Biomedical Imaging, Yale School of Medicine, New Haven, CT, USA
4 Department of Statistics & Data Science, Yale University, New Haven, CT, USA
2. Abstract
A deep learning model trained on some labeled data from a certain source domain generally performs poorly on data from different target domains due to domain shifts.
In this work, we achieve cross-modality domain adaptation, i.e. between CT and MRI images, via disentangled representations.
Domain adaptation is achieved in two steps:
- images from each domain are embedded into two spaces, a shared domain-invariant content space and a domain-specific style space.
- the representation in the content space is extracted to perform a task.
Moreover, our model achieved good generalization to joint-domain learning, in which unpaired data from different modalities are jointly learned to improve the segmentation performance on each individual modality.
3. Introduction
Real-world applications usually face varying visual domains.
The distribution differences between training and test data, i.e. domain shifts, can lead to significant
performance degradation.
Data collection and manual annotation for every new task and domain are time-consuming and expensive, especially for medical imaging, where data are limited and are collected from different scanners, protocols, sites, and modalities.
To solve this problem, domain adaptation algorithms look to build a model from a source data distribution that performs well on a different but related target data distribution. Wang, M., Deng, W.: Deep visual domain adaptation: A survey. Neurocomputing 312, 135{153 (2018)
While MRI acquisitions include more complex quantitative information than CT useful for liver segmentation, they are often less available clinically than CT images .
Thus, it would be helpful if we could learn a liver segmentation model from the more accessible CT data that also performs well on MRI images.
Contributions:
- This is the first to achieve unsupervised domain adaptation for segmentation via disentangled representations in the field of medical imaging.
- Our model decomposes images across domains into a domain-invariant content space, which preserves the anatomical information, and a domain-specific style space, which represents modality information.
4. Methodology
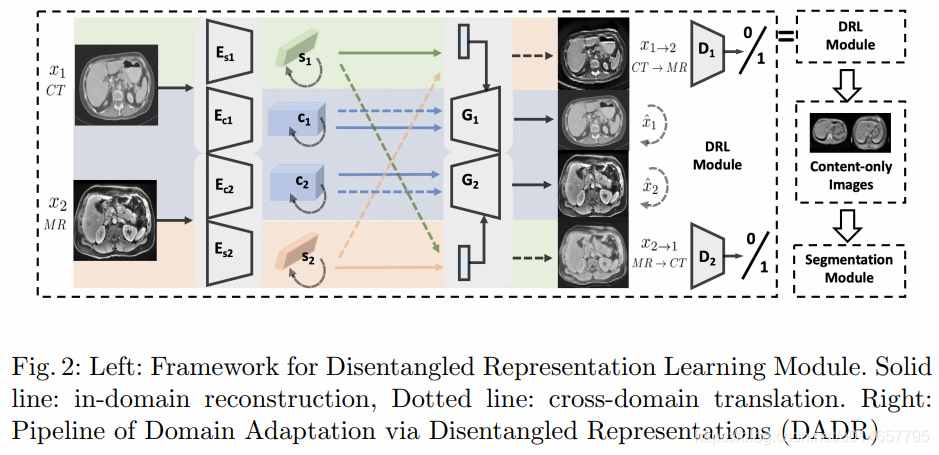
Our Domain Adaptation via Disentangled Representations (DADR) pipeline consists of two modules: Disentangled Representation Learning Module (DRLModule) and Segmentation Module (SegModule) (see Fig. 2). Of note, the DRL Module box in the DADR pipeline at the top is expanded in the left large box.
4.1 DRLModule
The module consists of two main components, a variational autoencoder (VAE) for reconstruction and a generative adversarial network (GAN) for adversarial training.
We train the VAE component for in-domain reconstruction, where reconstruction loss is minimized to encourage the encoders and generators to be inverses to each other.
The GAN component for cross-domain translation is trained to encourage the disentanglement of the latent space, decomposing it into content and style subspaces.
The overall loss function is defined as the weighted sum of the three components:
L
total
=
α
L
recon
+
β
L
a
d
v
+
γ
L
latent
L_{\text {total}}=\alpha L_{\text {recon}}+\beta L_{a d v}+\gamma L_{\text {latent}}
Ltotal=αLrecon+βLadv+γLlatent
- In-domain reconstruction,
L
recon
=
L
recon
1
+
L
recon
2
L_{\text {recon}}=L_{\text {recon}}^{1}+L_{\text {recon}}^{2}
Lrecon=Lrecon1+Lrecon2
L recon i = E x i ∼ X i ∥ G i ( E c i ( x i ) , E s i ( x i ) ) − x i ∥ 1 L_{\text {recon}}^{i}=\mathbb{E}_{x_{i} \sim X_{i}}\left\|G_{i}\left(E_{c i}\left(x_{i}\right), E_{s i}\left(x_{i}\right)\right)-x_{i}\right\|_{1} Lreconi=Exi∼Xi∥Gi(Eci(xi),Esi(xi))−xi∥1 - Cross-domain translation,
L
a
d
v
=
L
a
d
v
1
→
2
+
L
a
d
v
2
→
1
L_{a d v}=L_{a d v}^{1 \rightarrow 2}+L_{a d v}^{2 \rightarrow 1}
Ladv=Ladv1→2+Ladv2→1
L a d v 1 → 2 = E c 1 ∼ p ( c 1 ) , s 2 ∼ p ( s 2 ) [ log ( 1 − D 2 ( x 1 → 2 ) ) ] + E x 2 ∼ X 2 [ log ( D 2 ( x 2 ) ) ] L_{a d v}^{1 \rightarrow 2}=\mathbb{E}_{c_{1} \sim p\left(c_{1}\right), s_{2} \sim p\left(s_{2}\right)}\left[\log \left(1-D_{2}\left(x_{1 \rightarrow 2}\right)\right)\right]+\mathbb{E}_{x_{2} \sim X_{2}}\left[\log \left(D_{2}\left(x_{2}\right)\right)\right] Ladv1→2=Ec1∼p(c1),s2∼p(s2)[log(1−D2(x1→2))]+Ex2∼X2[log(D2(x2))] - Latent space reconstruction,
L
latent
=
L
recon
c
1
+
L
recon
s
1
+
L
recon
c
2
+
L
recon
s
2
L_{\text {latent}}=L_{\text {recon}}^{c_{1}}+L_{\text {recon}}^{s_{1}}+L_{\text {recon}}^{c_{2}}+L_{\text {recon}}^{s_{2}}
Llatent=Lreconc1+Lrecons1+Lreconc2+Lrecons2
L recon c 1 = ∥ E c 2 ( x 1 → 2 ) − c 1 ∥ 1 , L recon s 2 = ∥ E s 2 ( x 1 → 2 ) − s 2 ∥ 1 L_{\text {recon}}^{c_{1}}=\left\|E_{c_{2}}\left(x_{1 \rightarrow 2}\right)-c_{1}\right\|_{1}, L_{\text {recon}}^{s_{2}}=\left\|E_{s_{2}}\left(x_{1 \rightarrow 2}\right)-s_{2}\right\|_{1} Lreconc1=∥Ec2(x1→2)−c1∥1,Lrecons2=∥Es2(x1→2)−s2∥1
4.2 Domain Adaptation with Content-only Images
Once the disentangled representations are learned, content-only images can be reconstructed by using the content code c i c_{i} ci without style code s i s_{i} si.
4.3 Joint-domain Learning
Joint-domain learning aims to train a single model with data from both domains that works on both domains and outperforms models trained and tested separately on each domain.