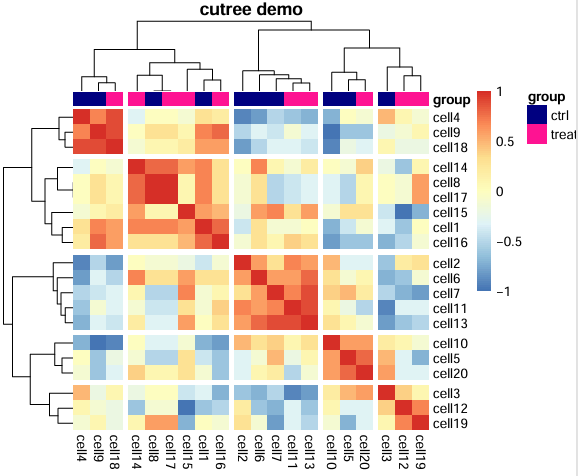
一个pheatmap的小例子,包含设置列注释,列注释颜色,聚类,聚类间隔,保存为pdf等。
- 加方框失败,可以后续使用Illustrator手工添加
效果图:
library(pheatmap)
set.seed(2025)
mat <- matrix(rnorm(100), nrow=20)
rownames(mat)=paste0("cell", 1:nrow(mat))
# 相关系数
mat.cor=cor( t(mat), method="spearman")
colnames(mat.cor)=rownames(mat)
# 列注释: 区分前10个ctrl组blue,后10个细胞treat组red
annotation_col = data.frame(
id=colnames(mat.cor),
group=c( rep("ctrl", 10), rep("treat", 10) ),
row.names=1
)
# 设置颜色
ann_colors = list(
group = c(ctrl = "navy", treat = "deeppink")
)
# 热图
library(grid)
#ComplexHeatmap::pheatmap(mat.cor,
p1=pheatmap::pheatmap(mat.cor,
#border_color = "white",
border_color = NA,
# 加方框:失败
#add_geom = "rectangles",
#rect_gp = gpar(fill = "transparent", col = "red", lwd = 3),
#rect_row = c(1, 5), rect_col = c(2, 7),
show_rownames = TRUE,
#gaps_row = c(1,3,7, 10), #适用于非聚类时
cutree_row=5, cutree_cols=5, #适用于聚类时
clustering_method = "ward.D2",
annotation_col = annotation_col, #annotation_row = annotation_row,
annotation_colors = ann_colors,
#color = c(colorRampPalette(colors = c("white","yellow"))(20),colorRampPalette(colors = c("yellow","firebrick3"))(20)),
show_colnames = TRUE, main="cutree demo")
#保存pdf
pdf(paste0("D://other//demo.heatmap.pdf"), width=6, height=5)
grid::grid.newpage()
grid::grid.draw(p1$gtable)
dev.off()